It’s been busy!
This week has been a busy one in the Williams lab.
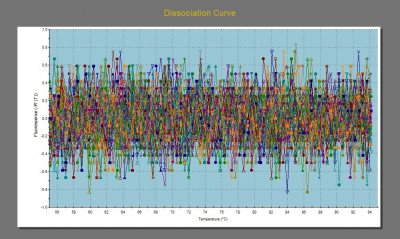
We had a bit more qPCR trouble in the beginning of the week. After running two successful plates on Monday, I ran a plate on Tuesday with unrecognizable results. To say the dissociation curves were messy is an understatement.
After a bit of troubleshooting and checking the instrument we realized that our replacement machine needed to be replaced. Until we get our new replacement we’re sharing the qPCR maching in Dana chemistry. We have to plan a little bit more carefully since we’re sharing with more people, but we’re lucky to have the second machine to hold us over.
On the bright side, I ran my last plate for the TL wild-type Nfe2 trials this afternoon! Once I learn how to use the new method of housekeeping normalization I can begin to analyze all the data thus far and then begin breeding again. We have Nfe2 knockdown fish that are just old enough to begin breeding. While we don’t have very many, using these fish will cut down on the workload immensely. Since we won’t need to inject, that not only saves time and skills practice, but also eliminates the need for the injection of control morpholino as a second control group to the wild-type TL trials. Essentially, that means three time points of DMSO and tBHQ that don’t need to be bred, collected, dosed, washed, and froze. We also won’t need to do the molecular work of RNA extraction, cDNA synthesis, and qPCR for 9 genes. I’d say it’s conservatively a month less of work, which brings the finish line closer and our goals much more attainable. I’m beginning to see the light at the end of the tunnel. I can’t wait to get my hand on the results of my last plate and get busy analyzing my data!